This HALRIC pilot project aims to identify new potential drug candidates and enzyme inhibitors using an innovative method of soaking small molecules into crystals of target proteins. The project focuses on two specific targets: SP3, an enzyme that degrades polyurethanes, and LAR, a protein involved in synapse formation and associated with psychiatric disorders.
In collaboration with the FragMAX team at the MAX IV Laboratory, the Thirup lab at Aarhus University (AU) will employ high-throughput X-ray diffraction to screen for fragments in crystals of the target molecules. This collaboration introduces a novel approach to the Thirup lab, utilizing the FragMax facility and the advanced Eiger2 CdTe detector at BioMAX for enhanced data collection. For this purpose, a researcher from AU will prepare high-purity samples of SP3 in collaboration with a group at the Technical University of Denmark (DTU).
The established protocol from the Thirup lab will be used to produce the required amounts of LAR. The project will utilize libraries containing a total of 1140 compounds for fragment soaking experiments, with data collection and processing supported by the BioMAX beamline team. Ultimately, the analysis of the screening results will be performed jointly by the research team, providing new insights into potential drug candidates and improving the functionality of plastic-degrading enzymes.
The project has two distinct goals using the Fragment-Based Drug Design approach:
1) LAR Protein: To map pockets and cavities on LAR, facilitating the design of drugs for treating psychiatric disorders.
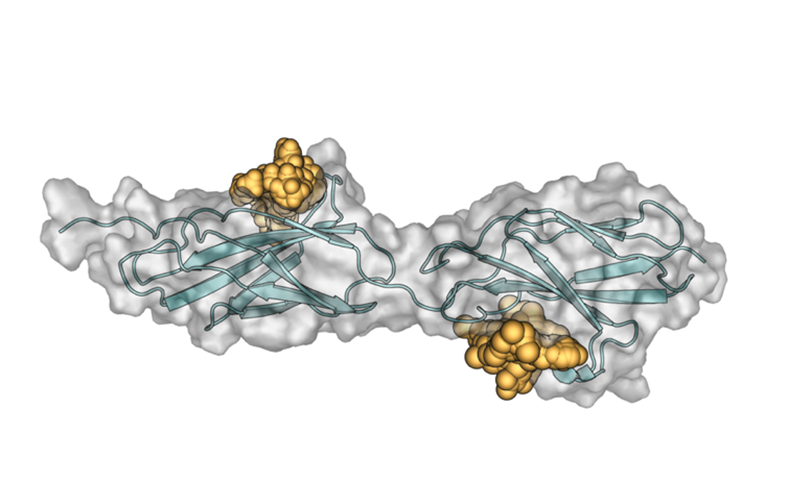
Figure1. Virtual drug screening results on LAR FN III domain 1-2 depicts small molecules (yellow spheres) targeting two shallow cavity/pocket on LAR.
2) SP3 Enzyme: To map the active and binding sites on SP3 and its homologs, understanding enzyme-plastic interactions. This knowledge will aid in protein engineering to enhance the enzyme’s activity in degrading polyurethane plastics, which are difficult to recycle.
 |
 |
 |
Figure2. Crystals structures of Polyurethane plastic degrading enzyme SP3 (green) and two thermostable homologs (gray and orange) determined by collaboration with J.P. Morth at DTU. Active site residues based on evolutionary conserved sequence is highlighted as sticks.
For further information about this HALRIC Pilot Project, please contact:
Deniz Bicer, Aarhus University
Deniz09@mbg.au.dk
